
This final assumption requires also that these references doesn’t change with the treatment or the course in question.
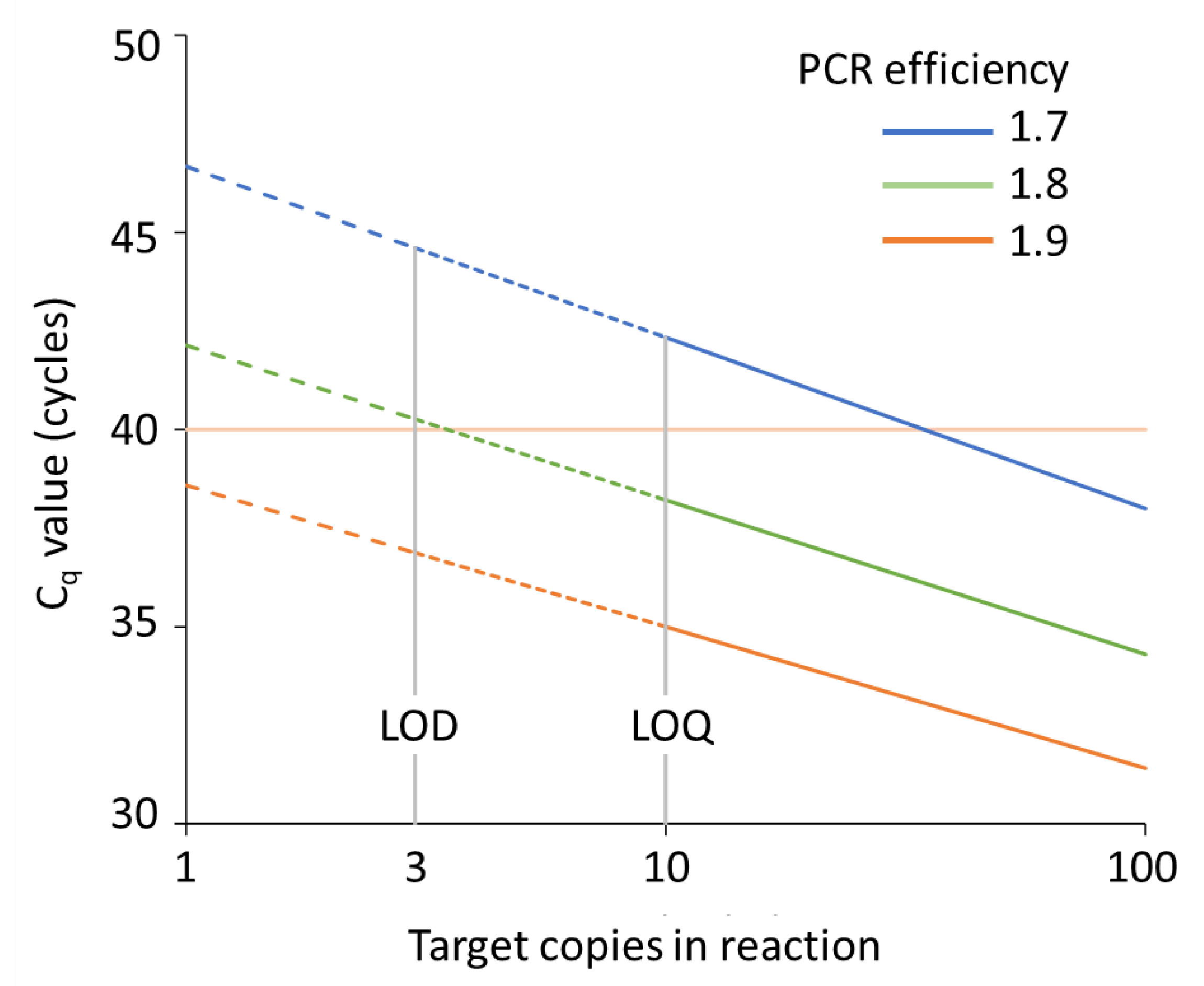
Another assumptions is that, the expression difference between two genes or two samples can be captured by subtracting one (gene or sample of interest) from another (reference). Meaning, at a certain threshold during the linear portion of the PCR reaction, the amount of the gene of the interest and the control double each cycle. And that this amplification efficiency is near perfect. The comparative \(C_T\) methods assume that the cDNA templates of the gene/s of interest as well as the control/reference gene have similar amplification efficiency. # locate and read dataįl <- system.file('extdata', 'ct3.csv', package = 'pcr')Īmount <- rep(c(1. The input ame is the \(C_T\) values of c-myc and GAPDH at different input amounts/dilutions. To assess the validity of this assumption, pcr_assess provides a method called efficiency. One of which is a perfect amplification efficiency of the PCR reaction.
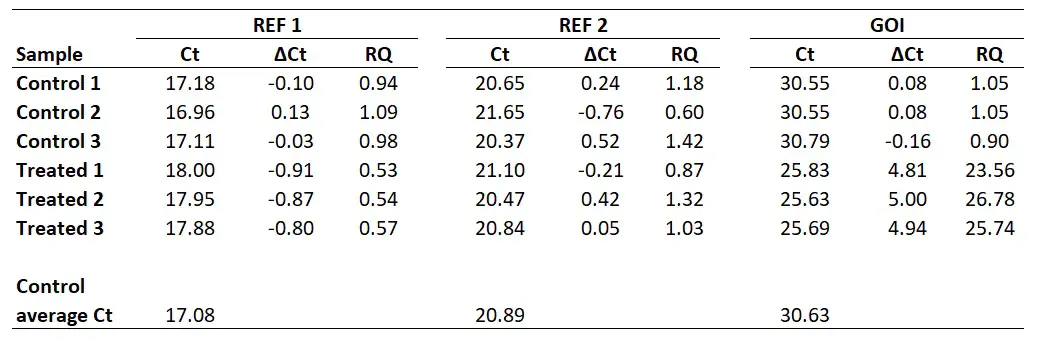
The previous analysis makes a few assumptions that will be explained later in this document. In addition, an error term and a lower and upper intervals are provided. Briefly, the input includes the \(C_T\) value of c-myc normalized to the control GAPDH, The calibrated value of c-myc in the kidney relative to the brain samples and the final relative_expression of c-myc. The output of pcr_analyze is explained in the documentation of the function ?pcr_analyze and the method it calls ?pcr_ddct as well as in a later scion of this document. # calculate all values and errors in one step Group_var <- rep(c('brain', 'kidney'), each = 6)

# default mode delta_delta_ctįl <- system.file('extdata', 'ct1.csv', package = 'pcr') pcr_analyze provides different methods, the default one that is used here is ‘delta_delta_ct’ applies the popular ( \(\Delta\Delta C_T\)) method.
#QPCR DATA ANALYSIS EXCEL CODE#
The following chunk of code locates a dataset of \(C_T\) values of two genes from 12 different samples and performs a quick analysis to obtain the expression of a target gene c-myc normalized by a control GAPDH in the Kidney samples relative to the brain samples.
#QPCR DATA ANALYSIS EXCEL INSTALL#
The development version of the package can be obtained through: # install package from githubĭevtools::install_github('MahShaaban/pcr') # load required libraries To install it, use: # install package CRAN


 0 kommentar(er)
0 kommentar(er)
